Can Two Species With the Same Genus Have Different Families
Abstruse
The genus Pseudomonas has gone through many taxonomic revisions over the past 100 years, going from a very big and diverse group of bacteria to a smaller, more refined and ordered list having specific backdrop. The relationship of the Pseudomonas genus to Azotobacter vinelandii is examined using iii genomic sequence-based methods. Starting time, using 16S rRNA trees, information technology is shown that A. vinelandii groups within the Pseudomonas shut to Pseudomonas aeruginosa. Genomes from other related organisms (Acinetobacter, Psychrobacter, and Cellvibrio) are exterior the Pseudomonas cluster. 2nd, pan genome family copse based on conserved factor families likewise show A. vinelandii to be more than closely related to Pseudomonas than other related organisms. Third, exhaustive BLAST comparisons demonstrate that the fraction of shared genes between A. vinelandii and Pseudomonas genomes is like to that of Pseudomonas species with each other. The results of these dissimilar methods point to a high similarity betwixt A. vinelandii and the Pseudomonas genus, suggesting that Azotobacter might actually be a Pseudomonas.
Introduction
Pseudomonas bacteria are naturally widespread in the environment. For example, the plant pathogen, Pseudomonas syringae has been linked to the environmental cycle of h2o as an ice nucleus in the clouds and is constitute in rain, snow, lakes, and plants [31]. Because of its abundance in the surround, the Pseudomonas genus was start characterized long agone, and over the past hundred years, information technology has gone through many taxonomic revisions. The number of organisms placed in the Pseudomonas group grew steadily over a period of 60 years. Notwithstanding, through refinement of defining criteria, many bacteria were moved to other genera over the adjacent l [24, 36, 42, 47].
Early studies based on rRNA–Dna hybridization postulated v RNA subdivisions in the genus, where rRNA grouping I, including the type species Pseudomonas aeruginosa, was named subsequently the genus as Pseudomonas [34]. Studies on the determination and comparison of 16S rRNA sequences of Pseudomonas species resulted in the clustering of Pseudomonas into two groups: P. aeruginosa and Pseudomonas fluorescens [32]. Afterwards, the extensive report of Anzai and collaborators on more than 100 Pseudomonas species based on 16S rRNA sequence comparison suggests seven clusters from the group of species of Pseudomonas sensu stricto, which besides agreed in some parts with Palleroni'due south study in 1973 [3]. Although it is nevertheless a widely accepted method, debates on the poor resolution of the phylogeny assay with rrs gene sequences lead to the idea of using other marker genes to characterize and allocate Pseudomonas, such every bit gryB, rpoD, oprI, oprF, and rpoB sequences [2, 8, 13, 57]. In some other study, ten housekeeping genes were used to assess the phylogeny of two,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. [16]. Other phenotypic methods, such as siderotyping, were also suggested for the classification of plant-associated Pseudomonas [thirty]. Pseudomonas sensu stricto (rRNA similarity group I) could be further divided into subgroups due to its considerable heterogeneity based on pathogenicity or pigment production [35].
The electric current status of the Pseudomonas genus today shows 202 species assigned to Pseudomonas on the Approved Lists of Bacterial Names, where the classification method depends on a combination of 16S rRNA, the analysis of the cellular fat acids, and differentiating classical physiological and biochemical tests [52]. The genus consists of a group of medically and biotechnologically important bacteria that are inhabitants of a broad range of niches including soil and water environments, in addition to plant and animal associations. Hence, they are well known for having enormous metabolic versatility [17, eighteen, 47]. They are not-sporulating, aerobic Gram-negative rods that are plant in biofilms or in planktonic forms. Most of the pathogenic members are related to plants, whereas several strains are pathogenic to animals [35].
Azotobacter vinelandii, in this context, is interesting because of its common metabolic characteristics with Pseudomonas. A nitrogen-fixing member of Gammaproteobacteria, A. vinelandii is institute mostly in soil environments where its nitrogen and energy metabolism is significant to agriculture. Many years ago, this organism was often used in biochemistry experiments for isolating enzymes during the kinetics studies which resulted in surprising yields and qualities [29]. It is a free-living obligate aerobe known for having the highest respiratory characteristics, merely it can still fix atmospheric nitrogen using a respiratory protection mechanism [23]. It also has distinct backdrop, such equally dramatic increase in chromosome numbers when reached at a stationary phase, formation of cysts under carbon depletion that helps the bacteria to resist aridity [41], where alginate is a structural component, and accumulation of poly-beta-hydroxybutyrate at the cease of the exponential growth as a carbon and energy source storage [48]. Although the Azotobacter genus has been studied over 100 years in various experiments, currently, there is only one complete genome sequence bachelor on NCBI GenBank database—A. vinelandii DJ [43]. There are no further ongoing projects listed for this genus, out of several thousand bacterial genome projects.
Azotobacter and Pseudomonas are members of the Pseudomonadaceae family. They both accept a significant genomic variety and genetic adaptability in a wide range of niches. However, numerous studies show that they share many biochemical metabolic pathways such as nitrogen fixation, alginate product, and respiratory mechanisms, and they are plant in similar environments [xi, 58]. It was long thought that Pseudomonas species (sensu stricto) exercise not have nitrogen fixation abilities; however, recently, it has been demonstrated that some Pseudomonas strains can gear up nitrogen and that their genes related to this machinery closely resemble that of A. vinelandii [39, 58, 59]. Another similarity is the alginate production in A. vinelandii, which is also a by-product in pathogenic P. aeruginosa infections in the lungs of cystic fibrosis patients [20]. However, other phenotypic characteristics of Pseudomonas have been shown to exist unlike from Azotobacter species, such as cell morphology and motility [35]. This suggests that the diversity in some phenotypic characteristics might be the effect of their adaptive backdrop since the two genera share the same set of core housekeeping genes or other conserved genes [59]. In this context, assay of 16S rRNA gene sequences by Rediers and collaborators showed that A. vinelandii was in the P. aeruginosa clade, sharing 96% identity with P. aeruginosa PAO1 strain. Owing to the low resolution in 16S rRNA sequences for these genomes, they conducted a phylogenetic analysis of 25 protein-coding genes, some of which were housekeeping genes. Phylogenetic trees generated with their dataset again revealed that A. vinelandii homologues are amassed within or close to the Pseudomonas group. The consensus tree out of these 25 topologies showed A. vinelandii phylogeny being closest to P. aeruginosa PAO1, terminal A. vinelandii to belong to the Pseudomonas genus [39]. Young and Park [59] use a broader arroyo to this idea, taking into account all the morphological differences, concluding that Azotobacter species tin be transferred to Pseudomonas along with a change in the criteria used for nomenclature.
In this article, we analyze the evolutionary relationships of the Pseudomonas genus to A. vinelandii, discussing whether or non this species is actually a Pseudomonas, using comparative genomic methods such as phylogeny copse, pan–core genome analysis, and protein BLAST across the whole genomes. Genomes from related genera—Acinetobacter, Psychrobacter, and Cellvibrio—were also used for the analysis to provide a better resolution. All genomes are therefore members of Pseudomonadales order, where Pseudomonas, Azotobacter, and Cellvibrio belong to the Pseudomonadaceae family unit and Acinetobacter and Psychrobacter are members of Moraxellaceae in the same order.
Materials and Methods
Gathering Genomes and Factor Annotation
All of the 29 genomes used in the analysis are complete sequences downloaded from GenBank [vi]. The list consists of A. vinelandii, 17 Pseudomonas species (including P. aeruginosa, Pseudomonas putida, Pseudomonas entomophila, Pseudomonas mendocina, P. fluorescens, P. syringae, and Pseudomonas stutzeri), 7 Acinetobacter, 3 Psychrobacter, and one Cellvibrio genome.
Gene annotations for the sequences were extracted from the GenBank files of each genome. Annotations from the GenBank files for the full genomes (including plasmids when present) were used. The properties of all the strains are listed in Table 1, and the colour coding in this table is used throughout the paper.
Phylogenetic Analysis—16S rRNA and Pan Genome Family unit Trees
Phylogenetic analysis was performed in two stages, i using 16S rRNA sequences and the other using the pan genome protein families. For the offset method, 16S rRNA sequences were predicted from the whole genome sequence using RNAmmer [26]. Unmarried and full sequences that have the highest scores from the RNAmmer predictions for each genome were aligned in CLUSTALW [27] plan, and with this alignment, the 16S rRNA tree was generated using a neighbor-joining method with 1,000 bootstrap resamplings and viewed with MEGA4 [50] (Fig. 1). An boosted tree was generated with partial 16S rRNA sequences of Azotobacter strains fractional sequences of Azotobacter strains—A. vinelandii IAM 15004 (type strain), Azotobacter tropicalis KBS, A. tropicalis RBS, Azotobacter nigricans IAM 15005 (type strain), Azotobacter armeniacus DSM 2284 (type strain), Azotobacter salinestris ATCC 49674(type strain), A. vinelandii ISSDS-384, Azotobacter beijerinckii ICMP 8673 (type strain), A. beijerinckii ICMP 4032, Azotobacter chroococcum IAM 12666(type strain), A. chroococcum ICMP 4031, Azomonas macrocytogenes ICMP 8674, Azorhizophilus paspali ATCC 23833, and Rhizobacter dauci H6—which are retrieved from the Ribosomal Database Project (RDP) [12], where the same procedure as above was performed, aligned with CLUSTALW, and viewed with MEGA4 [50] (come across Electronic supplementary textile (ESM) Fig. 1).
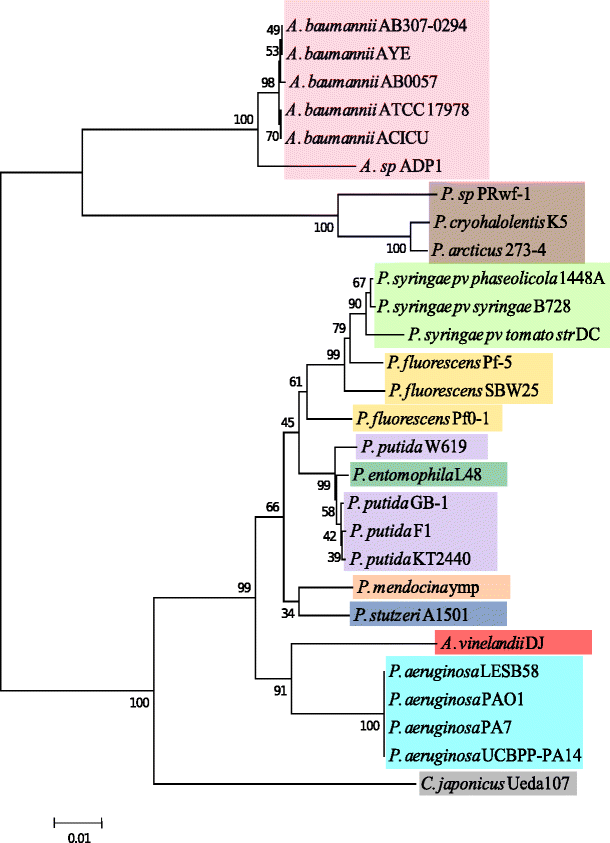
16S rRNA tree generated with the neighbor-joining method with 1,000 bootstrap resamplings. The tree shows the evolutionary relationships of A. vinelandii with the Pseudomonas genus and other Gammaproteobacteria based on their 16S rRNA sequences
For the 2nd method, the pan genome family tree (Fig. two) of all the genomes was generated using BLASTP similarity betwixt each proteome, as described in Snipen and Ussery'south method [45]. The "fifty/50 rule" was used to define the homology, meaning that a protein is causeless to be in the same family if 50% of its length shows 50% sequence identity with the reference protein [51]. Co-ordinate to this criterion, genes that have a significant striking to each other are considered to be in one gene family. In order to see the relations between the different gene families, a matrix is constructed containing the gene families in columns and the genomes in rows, assigning ane for the presence of that factor family in the corresponding genome and 0 otherwise. Manhattan distances are calculated from this matrix and hierarchical clustering is made. This tree shows the similarities based on the shared cistron families, excluding the factor families that are represented just in 1 genome (ORFans) [45].
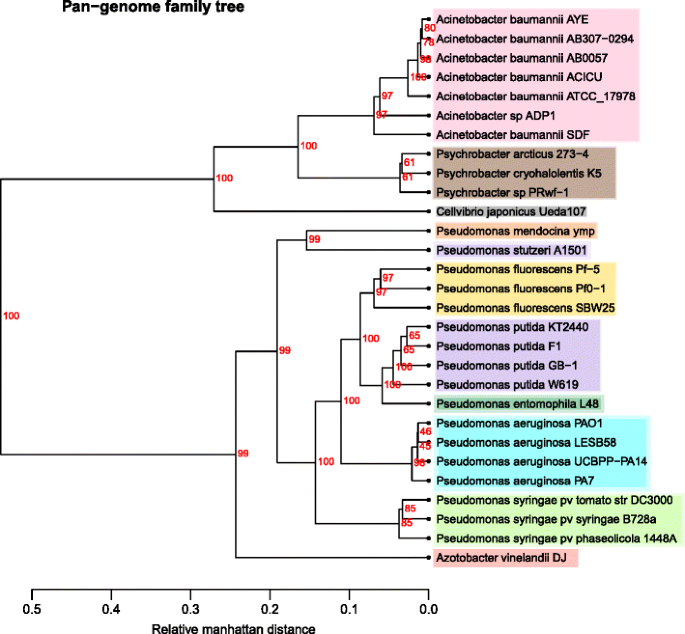
Pan genome family tree. The tree shows the phylogenetic relationships based on the gene families found in the pan genome, excluding the families found in simply one genome. Colour coding is again based on Table 1
Core and Pan Genome Analysis
In order to brand a pan–cadre genome analysis, BLASTP similarity analysis was used and a pan–core genome plot was generated based on the clustering from the pan genome family tree (Fig. 3). Pseudomonas genomes were plotted first, followed by the other genera. Azotobacter was added after P. stutzeri and so Acinetobacter, Psychrobacter, and, finally, Cellvibrio. The plot goes from the first to the final cavalcade in an information accumulative way. Each column shows the BLAST hit results against all the previous ones in terms of new genes, new cistron families, and pan and core genome sizes. The accumulative number of all factor families found (blue line in Fig. 3) increases as new genomes are added, leading to the pan genome in total, whereas the number of common cistron families found in all genomes (scarlet line in Fig. 3) decreases with genome addition, leading to the cadre genome at the end [53].
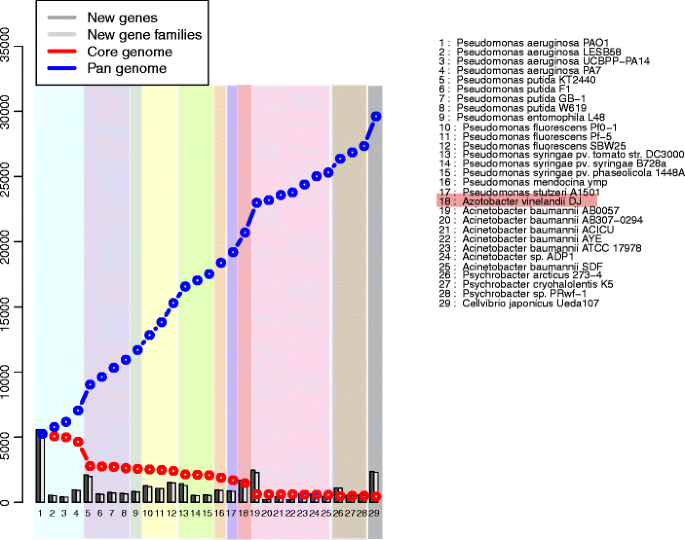
Core and pan genome plot of the Pseudomonas genomes. Azotobacter is highlighted. The number of pan genome for all strains used is 29,696 (dashed line on tiptop) and the core genome is reduced to 443 (dashed line beneath). The light purple colour indicates where P. putida is added, and the light blood-red column indicates where A. vinelandii is added
BLASTMatrix
In this method, the protein Smash identities were used to compare the proteome of each genome confronting all the other proteomes, pictured past a BLASTMatrix (Fig. iv), showing the fraction of genes shared between different genomes in the green cells [7]. The percentages in the cells are calculated by the number of factor families shared in two genomes over the union of the cistron families found in both. Similarity criterion was over again the fifty/fifty rule for the selection of significant hits and for the gene family unit assignment every bit in the pan genome family tree method. The term "homology" is as well used to bespeak the similarity based on shared cistron families between ii genomes. Protein BLAST results of a genome against itself (excluding self-hits) were used to define the protein homology within the same genome, every bit seen in the scarlet cells.
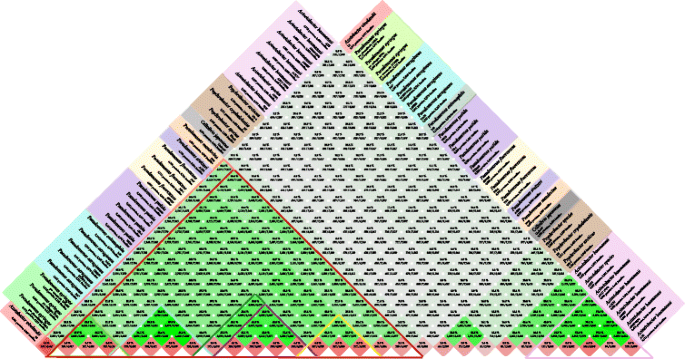
BLASTMatrix shows the homology between the pairs of genomes compared (green) and homology within each genome itself (ruby-red) based on the 50/50 rule
Results
16S rRNA Tree
According to the 16S rRNA phylogenetic tree in Fig. i, A. vinelandii has a close human relationship with Pseudomonas. The tree shows that the 16S rRNA sequence of this organism is very similar to P. aeruginosa 16S rRNA sequences, even more like than the other Pseudomonas species every bit mentioned before in Rediers et al. [39]. There are articulate clusters among the unlike strains of the same species, with few exceptions, and as expected, genomes from the genus Pseudomonas cluster together when compared with the other Pseudomonadales, Acinetobacter, and Psychrobacter, which are more distantly related. In general, the evolutionary relationships indicated in this tree are in agreement with the known biology of these organisms. Furthermore, alignment of sequences including more 16S rRNA sequences obtained from RDP as well shows the same clustering results for Azotobacter (encounter ESM Fig. 1). All the sequences from Azotobacter, Azorhizophilus, and Azomonas genera cluster close to the P. aeruginosa group, while other Pseudomonas species are more distant and Rhizobacter is an outgroup (notation that it was not specifically selected as an outgroup during the methods).
Pan Genome Family unit Tree
The pan genome family tree (Fig. two) compares the presence of gene families in each genome and measures the distances for the tree depending on the common factor families found between genomes, resulting in groups which share more factor families clustering together. As expected, the Pseudomonas genus groups together and has clear clusters for each species, this time with oft 100% bootstrap values in the main nodes for each species. Some positions of the genomes are changed, such as Cellvibrio japonicus and P. fluorescens, but, almost chiefly for this piece of work, the A. vinelandii now is in a different position on this tree. In this effigy, A. vinelandii does not group with any private Pseudomonas species but with all of the Pseudomonas clusters, still indicating that it shows a larger fraction of conserved protein families with Pseudomonads than the other Gammaproteobacteria.
Core and Pan Genome Assay
The core and pan genome analysis is another method that uses the proteomes of the genomes (Fig. 3). The plot shows the change in the number of gene families that are common to the compared genomes, the cadre genome, and the pan genome [51]. The bars bespeak the new genes and gene families compared with a BLASTP against the genomes that were previously added to the list. Hence, every new gene family is accounted for in the pan genome, which increases with the addition of each new genome (blue line in Fig. 3), whereas the size of the core genome is reduced (red line). The order of the genomes is related to the evolutionary distance seen on the pan genome tree.
The overall upshot of the core and pan genome plot shows that there are only 443 conserved core gene families found in 29 genomes. The cadre genome size for just the Pseudomonas genomes is 1,706, and after A. vinelandii is added, it is reduced by 231 cistron families, leading to 1,475 core gene families for the first xviii genomes. The pan genome size for all the strains adds upwards to 29,626 gene families. The increase in pan genome size after the add-on of A. vinelandii is 1,506 gene families, and there are roughly 1,700 genes that are designated as new. It is likewise seen that later the improver of Pseudomonas putida strains, the cadre genome for Pseudomonads has a steep drib with one,870 gene families and the pan genome has a sharp increase of 1,969 more gene families. There is also a big jump of 2,275 gene families in the pan genome after the addition of Acinetobacter species, where the core genome drops by 855 gene families.
BLASTMatrix
The BLASTMatrix shows the shared gene families between and within the compared genomes (Fig. 4). There is a loftier fraction of shared genes inside the same species, as denoted in darker green colors on the bottom of the matrix. The genus Pseudomonas tin can readily be distinguished, and information technology is highlighted with the night blood-red triangle. The results closely resemble the relations in the pan genome tree where Psychrobacter and Acinetobacter have a very depression homology with Pseudomonas species. Azotobacter on the other mitt shares every bit many gene families with Pseudomonas as some of the other members of Pseudomonadaceae. More specifically, it shares between 24% and 31% of its protein families with Pseudomonas; the maximum homology is seen with P. stutzeri with 31.2%, where P. stutzeri is homologous with other Pseudomonas between 28% and 34%. Too seen from the matrix is the homology within the species which is on average 79% for P. aeruginosa, 66% for P. putida, 49% for P. fluorescens, 65% for P. syringae, and 72% for Acinetobacter baumannii. In contrast, homology beyond various species inside Pseudomonas indicates a level between 30% and 50%.
Discussion
The results of using dissimilar comparative genomic methods to sympathize the phylogeny of Pseudomonas bespeak a considerably close evolutionary relationship with A. vinelandii. Both in the 16S rRNA tree and pan genome family tree, A. vinelandii is clustered together or within the Pseudomonas species. It is not equally clear whether or non A. vinelandii should be classified as being closest to P. aeruginosa. Although they are amassed together in the 16S rRNA tree, the low resolution of the 16S rRNA phylogenetic analysis has been noted by others [39, 59]. It does show, however, that they have a common ancestral rrs cistron that is closer to each other than to other species. The consequence of the 16S rRNA tree too shows that members of the Moraxellaceae family (Acinetobacter and Psychrobacter) are clustered together and Pseudomonodaceae (Pseudomonas, Azotobacter, and Cellvibrio) are on some other clade. In the pan genome family unit tree, on the other hand, clear clusters of each species can be seen. For case, P. fluorescens strains are in a grouping rather than separated as in the rRNA tree. Since this tree shows the distances of each group according to how many factor families they share, it is clear that A. vinelandii shares more gene families with the Pseudomonas grouping than it does with the other Pseudomonadales members used in this comparison, especially when compared with Cellvibrio, which is also another genus in the same family unit. Since the results are restricted to just ane Azotobacter genome, other supportive results are crucial in order to sympathize the true relationship.
The core and pan genome assay reveals the set of conserved gene families ("core") and the full number of gene families ("pan genome") for the set of sequenced genomes compared. The core genome refers to the thought of a backbone genome for organisms in the same genus; as more species from the same genus are added, the core is expected to approach a stable plateau in the same genus. The pan genome is, however, very flexible in size and, for some bacteria, can be quite big [53]. For instance, the pan genome of Pseudomonas is more than ten times larger than its cadre genome [25]. Effigy 3 shows that the Pseudomonas core genome size does not have a dramatic change with the improver of A. vinelandii. Although the pan genome has a slight increase, such a small change in the core genome size is not expected from an organism that is in a unlike genus. The abrupt decrease in the core genome when P. putida is added is due to its position on the list, being right afterward P. aeruginosa, which is clearly in another group in the phylogeny copse. However, subsequently eighteen genomes, the addition of a new genus (in this case, an Acinetobacter species) creates a big difference in the plot, reducing the cadre more than half in size and expanding the pan genome conspicuously by 10%, which is an obvious result of the improver of a different genus on the list.
The BLASTMatrix also strongly agrees with the pan genome family unit tree and pan–cadre genome plot as they are all comparing the gene families across the genomes. Among them, the BLASTMatrix provides more quantitative results on the similarities such as percentages or number of gene families. Co-ordinate to this, Azotobacter has a high fraction of shared protein families with Pseudomonas, mostly with P. stutzeri having 31%. They too share a like fraction of protein family levels with other Pseudomonas, with A. vinelandii having on boilerplate 25% homology and P. stutzeri with on average 30%. The relation between these two organisms might exist because of the similar functions that they have in their environment as they are both gratuitous-living, root-associated, nitrogen-fixing leaner [58]. However, phylogenetic analysis, based on 16S rRNA in this newspaper and in the other works besides based on housekeeping genes, suggests that A. vinelandii could have a closer evolutionary human relationship with P. aeruginosa. Taking into account that P. aeruginosa is the blazon species for Pseudomonas and that it shares a common evolutionary history on the conserved genes with A. vinelandii, all of these results strongly suggests that A. vinelandii has a Pseudomonas-like backbone including the conserved genes, while the diverseness among them is caused by adaptive strategies during their evolution which comes from the transfer of genetic material.
It should be noted that complete genomes were chosen in the dataset for the reliability of the methods. For many of the incomplete genomes in the database, 16S rRNA sequences are missing or fractional, and so as the many protein sequences. In a report where comparative analysis mostly relies on the Nail comparison on the proteomes, fractional or poorly annotated sequences cause artifacts in the search, which leads to unreliable conclusions. Another aspect from the taxonomical point of view is that in theory, a adept nomenclature method should be able to show the relations of organisms regardless of the number of data. Hence, having more Pseuodmonas genomes should not be the effecting factor in the discrimination of the organisms, specially at the genus level. On the other manus, having more Azotobacter genomes would be more reliable in terms of reducing the issue of sequencing, assembly, and annotation errors.
Concluding remarks
The increase in the availability of genome sequences of different organisms makes comparative genomic analysis a fundamental part of inquiry on evolutionary relations between organisms and the genetic footing of their diverseness [53]. This applies likewise to Pseudomonas. Looking at the comparative genomic assay of nucleotide and the coded protein sequences of Pseudomonas, we can easily see the distinction betwixt Pseudomonas species. This distinction is supported past the concrete and biochemical classification that has been established over the last 100 years.
Although there is only one Azotobacter genome available, general assumptions from these comparative analyses that rely on the extensively studied algorithms and tools are worth examining. Support from similar assay on the comparing of these genera is also taken into account. In conclusion, we suggest that A. vinelandii has a Pseudomonas-similar backbone genome where the core functions of these groups are same. There are various lines of evidence which atomic number 82 to this: the finding that A. vinelandii has approximately a third of the aforementioned protein families, clustering with the whole Pseudomonas clade on the pan genome family tree rather than being in the P. aeruginosa clade, and not causing a big drop on the core genome size. All three of these observations are consistent with the thought of having the same backbone, maybe the same origins, but different adaptations throughout their evolution. This leads united states to the question of the boundaries of new member assignments in the Pseudomonas genus. Where practice we draw the line? We propose that based on whole-genome assay, information technology is possible to meliorate differentiate members of a genus by looking at their core genome properties. The standards for nomenclature should be set using comparative methods on both Dna and protein levels. If this is the example, the Azotobacter can be assigned to the Pseudomonas genus. Perhaps, for future investigations, a detailed analysis of the cadre genes can be made and functionally categorized to see the background of their similarity.
References
-
Adams Md, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ et al (2008) Comparative genome sequence assay of multidrug-resistant Acinetobacter baumannii. J Bacteriol 190:8053–8064
-
Ait Tayeb L, Ageron E, Grimont F, Grimont PAD (2006) Molecular phylogeny of the genus Pseudomonas based on rpoB sequences and application for the identification of isolates. Res Microbiol 156:763–773
-
Anzai Y, Kim H, Park JY, Wakabayashi H, Oyaizu H (2000) Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int J Syst Evol Microbiol 50(Pt 4):1563–1589
-
Bakermans C, Tsapin AI, Souza-Egipsy Five, Gilichinsky DA, Nealson KH (2003) Reproduction and metabolism at −10°C of leaner isolated from Siberian permafrost. Environ Microbiol v:321–326
-
Barbe V, Vallenet D, Fonknechten N, Kreimeyer A, Oztas Southward, Labarre L et al (2004) Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res 32:5766–5779
-
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2008) GenBank. Nucleic Acids Res 36:D25–D30
-
Binnewies TT, Hallin PF, Staerfeldt HH, Ussery DW (2005) Genome Update: proteome comparisons. Microbiology 151:one–iv
-
Bodilis J, Hedde M, Orange N, Barray S (2006) OprF polymorphism as a marker of ecological niche in Pseudomonas. Environ Microbiol 8:1544–1551
-
Buell CR, Joardar 5, Lindeberg M, Selengut J, Paulsen It, Gwinn ML et al (2003) The consummate genome sequence of the Arabidopsis and tomato plant pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA 100:10181–10186
-
Chi CY, Lai CH, Fung CP, Wang JH (2005) Pseudomonas mendocina spondylodiscitis: a case written report and literature review. Scand J Infect Dis 37:950–953
-
Clementi F (1997) Alginate production by Azotobacter vinelandii. Crit Rev Biotechnol 17:327–361
-
Cole JR, Wang Q, Cardenas East, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145
-
De Vos D, Bouton C, Sarniguet A, De Vos P, Vauterin Chiliad, Cornelis P (1998) Sequence variety of the oprI gene, coding for major outer membrane lipoprotein I, among rRNA grouping I pseudomonads. J Bacteriol 180:6551–6556
-
DeBoy RT, Mongodin EF, Fouts DE, Tailford LE, Khouri H, Emerson JB et al (2008) Insights into institute cell wall degradation from the genome sequence of the soil bacterium Cellvibrio japonicus. J Bacteriol 190:5455–5463
-
Feil H, Feil WS, Chain P, Larimer F, DiBartolo G, Copeland A et al (2005) Comparison of the consummate genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato plant DC3000. Proc Natl Acad Sci United states of america 102:11064–11069
-
Frapolli One thousand, Défago Thou, Moënne-Loccoz Y (2007) Multilocus sequence analysis of biocontrol fluorescent Pseudomonas spp. producing the antifungal compound ii,4-diacetylphloroglucinol. Environ Microbiol 9:1939–1955
-
He J, Baldini RL, Déziel E, Saucier M, Zhang Q, Liberati NT et al (2004) The wide host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and creature virulence genes. Proc Natl Acad Sci United states 101:2530–2535
-
Høiby N, Ciofu O, Bjarnsholt T (2010) Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol 5:1663–1674
-
Iacono M, Villa Fifty, Fortini D, Bordoni R, Imperi F, Bonnal RJP et al (2008) Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II grouping. Antimicrob Agents Chemother 52:2616–2625
-
Jelsbak 50, Johansen HK, Frost AL, Thogersen R, Thomsen LE, Ciofu O et al (2007) Molecular epidemiology and dynamics of Pseudomonas aeruginosa populations in lungs of cystic fibrosis patients. Infect Immun 75:2214–2224
-
Joardar Five, Lindeberg K, Jackson RW, Selengut J, Dodson R, Brinkac LM et al (2005) Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals deviation amongst pathovars in genes involved in virulence and transposition. J Bacteriol 187:6488–6498
-
Kao CM, Liu JK, Chen YL, Chai CT, Chen SC (2005) Factors affecting the biodegradation of PCP by Pseudomonas mendocina NSYSU. J Risk Mater 124:68–73
-
Kennedy C, Bishop P (2005) Genetics of nitrogen fixation and related aspects of metabolism in species of Azotobacter: history and current condition. In: Klipp W, Masepohl B, Gallon JR, Newton WE (eds) Genetics and regulation of nitrogen fixation in free-living bacteria. Kluwer, Dordrecht, pp 27–52
-
Kersters Chiliad, Ludwig W, Vancanneyt M, Devos P, Gillis M, Schleifer KH (1996) Recent changes in the classification of the pseudomonads: an overview. Syst Appl Microbiol nineteen:465–477
-
Kiil K, Binnewies TT, Willenbrock H, Hansen SK, Yang Fifty, Jelsbak L et al (2008) Comparative genomics of Pseudomonas. In: Rehm BHA (ed) Pseudomonas. Wiley-VCH, Weinheim, pp 1–24
-
Lagesen Chiliad, Hallin P, Rødland EA, Staerfeldt HH, Rognes Tr, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108
-
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H et al (2007) Clustal W and Clustal 10 version ii.0. Bioinformatics 23:2947–2948
-
Lee DG, Urbach JM, Wu Thou, Liberati NT, Feinbaum RL, Miyata S et al (2006) Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol 7:R90
-
Lineweaver H, Burk D (1934) The determination of enzyme dissociation constants. J Am Chem Soc 56:658–666
-
Meyer JM, Gruffaz C, Fischer-LeSaux M (2008) Siderotyping, a straightforward tool to identify soil and plant-related pseudomonads. In: Nautiyal CS, Patrice D (eds) Molecular mechanisms of found and microbe coexistence. Springer, Berlin, pp 369–382
-
Morris CE, Sands DC, Vinatzer BA, Glaux C, Guilbaud C, Buffière A, Yan S, Dominguez H, Thompson BM (2008) The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J 2:321–334
-
Moore ERB, Mau M, Arnscheidt A, Boettger EC, Hutson RA, Collins Physician, Van de Peer Y, De Wachter R, Timmis KN (1996) The conclusion and comparison of the 16S rRNA gene sequences of species of the genus Pseudomonas (sensu stricto) and interpretation of the natural intrageneric human relationship. Syst Appl Microbiol 19:478–492
-
Nelson KE, Weinel C, Paulsen It, Dodson RJ, Hilbert H, Martins dos Santos VAP et al (2002) Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol 4:799–808
-
Palleroni NJ, Kunisawa R, Contopoulou R, Doudoroff M (1973) Nucleic acid homologies in the genus Pseudomonas. Int J Syst Bacteriol 23:333–339
-
Palleroni NJ (2005) Genus I. Pseudomonas Migula 1894, 237AL. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, Boone DR, Vos P et al (eds) Bergey'south manual of systematic bacteriology. Springer, Boston
-
Palleroni NJ (2008) The route to the taxonomy of Pseudomonas. In: Cornelis P (ed) Pseudomonas. Genomics and molecular biological science. CaisterAcademic, Norfolk, pp one–eighteen
-
Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GSA, Mavrodi DV et al (2005) Consummate genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol 23:873–878
-
Phoenix P, Keane A, Patel A, Bergeron H, Ghoshal S, Lau PCK (2003) Label of a new solvent-responsive gene locus in Pseudomonas putida F1 and its functionalization as a versatile biosensor. Environ Microbiol 5:1309–1327
-
Rediers H, Vanderleyden J, de Mot R (2004) Azotobacter vinelandii: a Pseudomonas in disguise? Microbiology 150:1117–1119
-
Roy PH, Tetu SG, Larouche A, Elbourne Fifty, Tremblay S, Ren Q, Dodson R, Harkins D, Shay R, Watkins Grand, Mahamoud Y, Paulsen Information technology (2010) Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS One 5(ane):e8842
-
Sadoff HL (1975) Encystment and germination in Azotobacter vinelandii. Bacteriol Rev 39:516–539
-
Schleifer KH (2009) Classification of Leaner and Archaea: past, present and futurity. Syst Appl Microbiol 32:533–542
-
Setubal JC, dos Santos P, Goldman BS, Ertesvåg H, Espin G, Rubio LM et al (2009) Genome sequence of Azotobacter vinelandii, an obligate aerobe specialized to back up diverse anaerobic metabolic processes. J Bacteriol 191:4534–4545
-
Silby MW, Cerdeño-Tárraga AM, Vernikos GS, Giddens SR, Jackson RW, Preston GM et al (2009) Genomic and genetic analyses of diversity and found interactions of Pseudomonas fluorescens. Genome Biol 10:R51
-
Snipen L, Ussery D (2010) Pan-genome trees. Stand Genomic Sci 2:135–141
-
Smith MG, Gianoulis TA, Pukatzki Due south, Mekalanos JJ, Ornston LN, Gerstein K, Snyder Thousand (2007) New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev 21:601–614
-
Stanier RY, Palleroni NJ, Doudoroff M (1966) The aerobic pseudomonads: a taxonomic study. J Gen Microbiol 43:159–271
-
Stevenson LH, Socolofsky MD (1966) Cyst formation and poly-{beta}-hydroxybutyric acid aggregating in Azotobacter. J Bacteriol 91:304–310
-
Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ et al (2000) Consummate genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964
-
Tamura K, Dudley J, Nei Yard, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Assay (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
-
Tettelin H, Masignani Five, Cieslewicz MJ, Donati C, Medini D et al (2005) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome". Proc Natl Acad Sci Usa 102:13,950–13,955
-
Tindall BJ, Kampfer P, Euzeby JP, Oren A (2006) Valid publication of names of prokaryotes according to the rules of nomenclature: past history and current practise. Int J Syst Evol Microbiol 56:2715–2720
-
Ussery DW, Wassenaar TM, Borini Southward (2009) Calculating for comparative microbial genomics. Springer, London
-
Vallenet D, Nordmann P, Barbe Vr, Poirel L, Mangenot Southward, Bataille Eastward et al (2008) Comparative analysis of Acinetobacters: 3 genomes for 3 lifestyles. PLoS I 3:e1805
-
Vodovar North, Vallenet D, Cruveiller Sp, Rouy Z, Barbe Vr, Acosta C et al (2006) Complete genome sequence of the entomopathogenic and metabolically versatile soil bacterium Pseudomonas entomophila. Nat Biotechnol 24:673–679
-
Winstanley C, Langille MGI, Fothergill JL, Kukavica-Ibrulj I, Paradis-Bleau C, Sanschagrin F et al (2009) Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool epidemic strain of Pseudomonas aeruginosa. Genome Res 19:12–23
-
Yamamoto S, Kasai H, Arnold DL, Jackson RW, Vivian A, Harayama S (2000) Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 146:2385–2394
-
Yan Y, Yang J, Dou Y, Chen K, Ping Southward, Peng J et al (2008) Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proc Natl Acad Sci United states 105:7564–7569
-
Young JM, Park DC (2007) Probable synonymy of the nitrogen-fixing genus Azotobacter and the genus Pseudomonas. Int J Syst Evol Microbiol 57:2894–2901
Acknowledgments
Nosotros would like to give thanks Peter Fisher Hallin, PhD, from Center for Biological Sequence Assay(CBS), Department of Systems Biology, The Technical University of Denmark, currently in Novozymes; Karin Lagesen from CBS, DTU and Centre for Molecular Biology & Neuroscience and Institute of Medical Microbiology, University of Oslo; Lars Snipen from Biostatistics, Dept. Chemical science, Biotech., and Food Sciences, Norwegian University of Life, for help in providing authentic and useful tools for this assay, and Colleen Ussery for carefully proofreading and editing the manuscript. This enquiry was supported in part by a grant 09-067103/DSF from the Danish Quango for Strategic Inquiry.
Open Access
This article is distributed under the terms of the Artistic Eatables Attribution Noncommercial License which permits any noncommercial utilise, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author data
Affiliations
Corresponding writer
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Admission This is an open access article distributed nether the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/past-nc/2.0), which permits any noncommercial utilise, distribution, and reproduction in whatever medium, provided the original writer(due south) and source are credited.
Reprints and Permissions
About this article
Cite this article
Özen, A.I., Ussery, D.Due west. Defining the Pseudomonas Genus: Where Do We Describe the Line with Azotobacter?. Microb Ecol 63, 239–248 (2012). https://doi.org/10.1007/s00248-011-9914-8
-
Received:
-
Accepted:
-
Published:
-
Issue Engagement:
-
DOI : https://doi.org/ten.1007/s00248-011-9914-8
Keywords
- Pseudomonas
- Acinetobacter
- Azotobacter
- Cadre Genome
- Pseudomonadales
thrasheroultiventy.blogspot.com
Source: https://link.springer.com/article/10.1007/s00248-011-9914-8
0 Response to "Can Two Species With the Same Genus Have Different Families"
Post a Comment